osteomiR® – miRNA Biomarkers | TW-KT-011-OT
-
Assay time
Sample to data in 8 hours with 2 hours hands-on time
-
Sample volume
200 µl / sample
-
Sample type
Serum, EDTA plasma
-
Method
RT-qPCR, relative quantification without standard curve
-
Sensitivity
10 copies / µl
-
Regulatory status
Research use only.
-
Specificity
19 selected bone-related microRNAs with characterized contributions to bone disease
-
Use
Research use only.
Product Overview
TAmiRNA have identified 19 microRNAs in human serum which
» are associated to bone microstructure and histomorphometry
» are regulated in patients with osteoporotic fractures
» are BMD-independent risk factors for osteoporotic fractures
» regulate bone formation and resorption via multiple pathways
» are novel biomarkers for bone diseases
» can easily be detected in human serum and plasma samples.
The osteomiR® kit has been developed to standardize the quantification of these selected microRNA biomarker candidates for fracture-risk in postmenopausal and diabetic osteoporosis. The osteomiR® kit is intended for research-use only, not for diagnosis, prevention or treatment of a disease. The clinical utility of the osteomiR® kit is currently investigated in clinical trials.
The osteomiR® kit provides users with a highly standardized method to determine the levels of the 19 informative microRNAs in human serum samples. It alleviates the task of selecting and optimizing analytical methods, data pre-processing and data normalization. It provides standardized serum concentrations for microRNAs with a known association to bone cell function, bone remodelling and fracture risk.
Principle of the osteomiR® RNA Extraction
The Serum/Plasma RNA extraction kit enables the isolation of miRNA, from a minimum of 200 μl of sample. The phenol-free protocol uses spin column technology without the need for a vacuum pump. It allows analysis of extracellular vesicle RNA through lysis of the vesicles. The kit is designed to isolate high quality microRNA in amounts sufficient for qPCR analysis using the osteomiR® kit.
The workflow consists of 5 simple steps:
1. Lysis of biofluid components
2. Precipitation and removal of proteins
3. Precipitation of RNA using isopropanol and column loading
4. Washing
5. Elution
In the first part of the RNA isolation process, membranized particles/cells are lysed using the provided lysis solution. Proteins are precipitated using the precipitation solution and the supernatant (including RNA) is mixed with isopropanol for precipitation. This solution is loaded onto a spin-column, where a resin binds RNA in a manner that depends on ionic concentrations. Thus, microRNA will bind to the column, while the residual proteins will be removed in the flow-through or retained on the top of the resin. The bound microRNA is then washed with the provided wash solutions in order to remove any remaining impurities, and the purified microRNA is eluted with RNase free water.
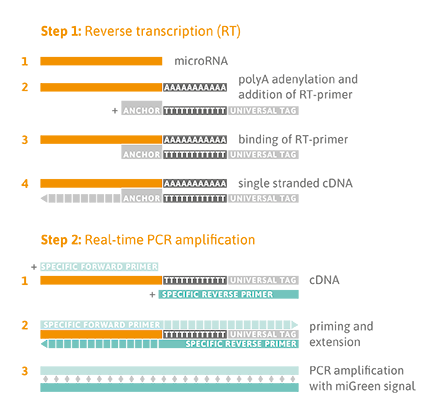
Reverse Transcription Quantitative PCR Detection
A universal reverse transcription (RT) reaction is performed, as shown in the Figure below (Step 1), which means that all microRNA species are converted into complementary DNA (cDNA) at the same time. This enables parallel quantitative PCR (qPCR) detection of different microRNA sequences in one cDNA sample using the osteomiR® plate. Universal RT is achieved by first adding a poly-A tail to the mature microRNA template (Step 1). Complementary cDNA is synthesized using a poly-T-primer with a 3’ degenerate anchor and a 5’ universal tag. During qPCR, the cDNA is then amplified using microRNA-specific and LNA™-enhanced forward and reverse primers using miGreen for detection of double-stranded DNA (Step 2).
LNA™ Technology
Locked Nucleic Acids (LNA™) are chemically modified nucleotides, which offer substantially increased affinity for its complementary strand, compared to traditional DNA or RNA oligonucleotides. This results in unprecedented sensitivity and specificity and makes LNA™ oligonucleotides ideal for the detection of microRNAs, due to their short length and varying content of G-C and A-T bases. Without LNA™, the heterogeneous hybridization properties could result in unspecific and low efficient primer binding and compromise data quality.
Kit Components
|
Contents |
Description |
Quantity |
|
qPCR plates |
Primer coated 96 or 384 well qPCR plates, 19 osteomiRs® and 5 controls/sample per plate; Storage: -20°C |
48 samples |
|
Serum/Plasma RNA extraction Kit |
Storage: RT |
48 samples |
|
osteomiR™ chemistry |
Spike-Ins, RT chemistry, miGreen Mix; Storage: -20°C; |
48 samples |
|
SOFTWARE |
osteomiR® software included to obtain normalized data and a fracture-risk score |
The osteomiR® chemistry will be shipped on dry ice and must be stored at -20°C. The osteomiR® qPCR plates will be shipped at RT and must be stored at -20°C. The Serum/Plasma RNA extraction kit is shipped at ambient temperature. Store the spin columns immediately at 2-8°C. Under these conditions, all components are stable until the expiry date on the package or vial.
qPCR Cycler Compatibility
All osteomiR® kits come with the components listed above and can be used for up to 48 samples. However, depending on your qPCR cycler select one of the following kits:
| Product number | Product | qPCR cycler compatibility |
| KT-011-OT 96A | osteomiR® 96-well | ABI, Bio-Rad, Eppendorf, Stratagene, Takara |
| KT-011-OT 96C | osteomiR® 96-well | ABI (fast block) |
| KT-011-OT 96D | osteomiR® 96-well | Bio-Rad, Stratagene, DNA Engine |
| KT-011-OT 96F | osteomiR® 96-well | Roche LC480 I and II |
| KT-011-OT 384E | osteomiR® 384-well | ABI, Bio-Rad CFX 384™ |
| KT-011-OT 384G | osteomiR® 384-well | Roche LC480 I and II |
Choice of Biofluid
TAmiRNA has used serum throughout its clinical development program for the discovery of microRNA biomarkers for bone quality. Therefore, we recommend to use serum for any experimental study using the osteomiR® kit. However, the kit has been shown to enable proper detection of osteomiRs also in EDTA-plasma samples.
Pre-analytical Standardization
Conditions during serum or plasma processing might affect the detection of microRNAs using qPCR. Therefore, we strongly recommend to standardize protocols for pre-analytical processing and serum collection. The following points should be incorporated in the pre-analytical study protocols:
- Patient variation: ensure overnight fasting prior to blood collection. Circadian rhythm, activity and diet are known to influence the levels of circulating microRNA content in patients.
- Use standardized needles and serum collection tubes. We recommend to use 21 gauge needles for blood collection. Ensure that only the specified blood collection materials are used during the entire study.
- Blood collection must be performed by a person that is well trained and familiar with the study protocol. Gloves must be worn at all times when handling specimens. This includes amongst others removal of the rubber stopper from the blood tubes, centrifugation, pipetting, disposal of contaminated tubes, and clean-up of any spills. Tubes, needles, and pipets must be properly disposed of in biohazard containers, in
accordance with institutional requirements. - Universal precautions and OSHA (Occupational Safety and Health Administration) and institutional requirements (http://www.osha.gov/SLTC/biologicalagents/index.html) should be followed, including gloves, eye protection or working in a biosafety cabinet for blood processing.
- Coagulation and centrifugation protocols for obtaining serum from whole blood must be standardized and followed strictly (see “Serum Collection” for more details).
- Hemolysis (visible as red-colored biofluid) must be recorded for all samples used (see “Quality Control” for more details on how to detect hemolysis in your samples).
- After thawing of collected serum samples, ensure that samples are kept cool (on ice or 4°C) at all times and avoid frequent freeze-thaw cycles. Low temperature is essential for RNA stability and sample matrix.
- For handling of serum as well as RNA/DNA we strongly advise to use RNase-free filter tips and nuclease-free microcentrifuge tubes with optimized surface properties to prevent adsorption of nucleic acids (“low binding”).
Storage and Stability of Serum, RNA and DNA
All samples should be stored in nuclease-free plastic tubes with minimized absorption rates for nucleic-acids “low binding”. To avoid freeze-thaw cycles the generation of aliquots of serum samples is recommended. We recommend aliquot volumes of 225 µl serum.
Serum, and RNA samples must be stored at -80°C for long term storage and kept on ice at all times during working procedures.
TAmiRNA has successfully used serum samples for microRNA analysis, which have been stored at -80°C for 15 years. In case the kit is intended to be used for serum samples that are older than 15 years, we recommend to conduct a feasibility study to assess the detection rates of microRNAs compared to fresh samples.
Total RNA samples should be stored in nuclease-free low-binding tubes for not more than 6 months prior to analysis.
Undiluted cDNA must be stored at -20°C and can be used for up to 5 weeks after initial storage. Avoid more than 5 freeze/thaw cycles of frozen RNA and undiluted cDNA samples.
Working with RNA
RNases are very stable and robust enzymes that degrade RNA. Before working with RNA, it is recommended to create an RNase-free environment following the precautions below:
- The RNase-free working environment should be located away from microbiological work stations.
- Clean, disposable gloves should be worn at all times when handling reagents, samples, pipettes, disposable tubes, etc.
- It is recommended that gloves are changed frequently to avoid contamination.
- Designated solutions, tips, tubes, lab coats, pipettes, etc. should be prepared for RNA
work only. - All solutions that will be used should be prepared using molecular biology grade nuclease-free water.
- Clean all surfaces with commercially available RNase decontamination solutions.
- When working with purified RNA samples, ensure that they remain on ice.
- Spin down all reaction and sample tubes before opening.
- Use filter barrier pipette tips to avoid aerosol-mediated contamination.
It is recommended to establish and maintain designated areas for PCR setup, PCR amplification, and DNA detection of PCR products, due to the risk of contaminating reagents and mastermixes with amplified DNA. The isolation of RNA and the reaction steps preceeding real-time PCR should be performed in rooms or areas, which are separated from areas where PCR experiments are performed in order to avoid contamination with amplified DNA. Use separate clean lab coats for RNA sample preparation, cDNA synthesis and when setting up PCR reactions or handling PCR products. Avoid bringing and opening tubes with amplified PCR products into the PCR setup area.
Quality Control
Synthetic Spike-In Controls
In general, spike-in controls are used to monitor the efficiency and correct result of every work step in the experiment. They can be used to identify outliers due to the presence of inhibiting factors or incorrect handling.
Uniform Cq-values obtained for the spike-ins demonstrate successful and homogenous RNA isolation, reverse transcription and qPCR for the samples. Synthetic spike-ins do not reveal the RNA content and quality in the biological sample.
UniSp4 – The synthetic RNA spike-in “UniSp4” is added to the sample during RNA extraction. It is used to monitor RNA extraction efficiency.
Cel-miR-39 – The synthetic RNA Spike-In “cel-miR-39” is added to the extracted RNA during reverse transcription into cDNA. It is used as a control for reverse transcription efficiency. It shares the natural microRNA sequence from C. elegans, which is not found in mammalian species. Reverse transcription efficiency is known to introduce the highest technical variance to RT-qPCR data
UniSp3 – The synthetic DNA UniSp3 is present at a fixed position on every osteomiR® test plate. It is used to monitor PCR efficiency and to detect the presence of PCR inhibitors in samples.
The results obtained for all three spike-ins should be carefully analyzed using the osteomiR® software app. It should be used to identify potential outliers, and to exclude samples from subsequent normalization and statistical analysis. Spike-ins can be used for calibration of Cq-data of informative microRNAs to remove technical variance.
Hemolysis
Hemolysis can be a major cause of variation in serum/plasma microRNA levels due to contamination with cellular RNA.
The presence of hemolysis should be assessed visually for each sample. In addition, hemolysis can be assessed using the hemolysis-index, which is based on the relative expression of miR-451a-5p compared to miR 23a-3p. An increase in miR-451a-5p relative to miR-23a-3p indicates the presence of hemolysis in human serum or plasma samples. The osteomiR® App will automatically calculate and report the Hemolysis Index.
Another option to determine hemolysis is the measurement of the absorbance peak of free haemoglobin by assessing free haemoglobin using a spectrophotometer such as NanoDrop™. Human serum or plasma samples are classified as being hemolyzed if the absorption at 414 nm is exceeding 0.2. However, the presence of small amounts of cellular contamination in serum or plasma samples is not readily detectable by visual or spectrophotometric means.
RNA Yield
Determination of RNA yield from 200 µl serum is not possible by optical spectrophotometry or NanoDrop™ due to the lack in sensitivity of the method. We therefore recommend to assess RNA yield and extraction efficiency using synthetic spike-in controls. In addition, the entire analytical protocol precisely specifies to fluid volumes throughout the entire workflow (see “Protocol”). The osteomiR® kit uses 200 µl serum for RNA extraction and 2 µl RNA for reverse transcription into cDNA. This is the optimum sample input in order to avoid inhibition of the reverse transcription reaction due to inhibitors that are co-extracted with total RNA. Excess amounts of total RNA in the reverse transcription reaction have been shown to lead to a non-linear quantification of microRNAs as well as a poor call rate.
Serum Collection
The osteomiR® Workflow requires 200 μl serum.
Serum collection is ideally performed after overnight fasting in the morning hours between 8 am and 10 am. This can reduce biological variance in miRNA levels due to activity and diet. We recommend the use of 21 gauge needles and red top vacutainer tubes (BD vacutainer®) for blood collection. Filled collection tubes should sit upright after the blood is drawn at room temperature for a minimum of 30 to a maximum of 60 minutes for the clot to form. The red top tubes do not have to be full to be used.
Centrifuge the blood sample at 2 500 g for 10 minutes at room temperature in a horizontal rotor (swing-out head). If the blood is not centrifuged immediately after the clotting time (30 to 60 minutes at room temperature), the tubes should be refrigerated (4ºC) for no longer than 4 hours. After centrifuging, the clot is located at the bottom of the tube, and the serum is on top of the clot. Use a clean pipette and nuclease-free filter tips to transfer the serum (recommendation: do not pour!). Pipette serum into the labeled nuclease-free (1.5 ml) tubes, filling the vials in sequential order. Aliquot volume is recommended to be 225 μl, so that 200 μl can be safely used for RNA extraction. Close the caps on the vials tightly. This process should be completed within 1 hour of centrifugation.
Check that all aliquot vial caps are secure and that all vials are labeled. Place all aliquots upright in a specimen box or rack in an -80ºC or colder freezer. All specimens should remain at -80ºC or colder prior to analysis or shipping. The sample aliquots should not be thawed prior to analysis or shipping.
Protocol
The RNA extraction has been standardized to a volume of 200 µl serum as starting material to ensure high RNA yield and prevent inhibition of downstream PCR applications.
If less than 200 µL serum is available we recommend to fill available serum up to 200 µl total volume using RNase-free water. Keep in mind that lower sample input might lead to a reduced sensitivity and yield.
RNA Extraction
Important points before starting
- Lysis Buffer and Wash Buffer 1 may form a precipitate upon storage. If necessary, redissolve by warming and then place at room temperature.
- Equilibrate buffers at room temperature before starting the protocol.
- Prepare 80% ethanol for Step 9c.
- All steps should be performed at room temperature. Work quickly!
This protocol is designed for human serum.
| 1. | Preparation of Wash Buffer 1 and Wash Buffer 2 First time use only |
Add 2 volumes of >99 % ethanol to Wash Buffer 1 and 4 volumes of >99 % ethanol to Wash Buffer 2. Note: The label on the bottle has a box that should be checked to indicate that the ethanol has been added. |
| 2. | Reconstitution of lyophilized spike-in controls (UniSp4 and cel-miR-39). First time use only |
1. Spin down vials before use by centrifugation at 3,000 g for 30 sec at room temperature. 2. Resuspend the spike-ins by adding 80 μl nuclease-free water. 3. Mix by vortexing and spin down. Store on ice for 20 min. 4. Mix by vortexing again and aliquot in low bind tubes (20 μl aliquots are recommended) 5. Store at -20°C. |
| 3. | Thaw serum sample | After thawing on room temperature, centrifuge the samples at 12,000 g for 5 min at 4°C to pellet any debris and insoluble components and to reduce effect of inhibitors/nucleases. Thaw glycogen on RT and store on ice. |
| 4. | Serum lysis | Transfer exactly 200 µl serum to a new 1.5 mL tube. If using less than 200 µl, fill up to 200 µl with RNase-free water. Ensure equal volumes of all samples. Add 1 µl UniSp4 to 60 µl Lysis Buffer and mix it with the 200 µl serum. Vortex for 5 sec and incubate for 3 min at room temperature. For multiple samples prepare a mastermix including 1 extra Rxn. Important note: The UniSp4 spike-in must be mixed with the lysis buffer before mixing with the sample – if added directly to the sample it will be rapidly degraded. |
| 5. | Protein precipitation | Add 20 µl of Protein Precipitation Buffer. Vortex for >20 sec and incubate for 3 min at room temperature. When processing multiple samples vortex immediately after addition of Protein Precipitation Buffer. The solution should become a milky suspension. Centrifuge for 3 min at 12,000 g at room temperature. |
| 6. | Transfer supernatant | Transfer exactly 200 µl of the clear supernatant (aqueous phase) into a new collection tube (1,5 ml, with lid) and add 2 µl glycogen (5 mg/ml). Vortex and spin down. |
| 7. | Adjust binding conditions | Add 200 µl isopropanol. Vortex for 5 sec. |
| 8. | Load column | Place a Spin Column in a collection tube and load the entire sample onto the column. Centrifuge for 30 sec at 8,000 g at room temperature. Discard flow-through and place column back into the collection tube. |
| 9a. | Wash and dry | Add 700 µl Wash Solution Wash Buffer 1 to the spin column. Centrifuge for 15 sec at 8,000 g at room temperature. Discard flow-through and place column back into the collection tube. |
| 9b. | Wash and dry | Add 500 μl Wash Solution Wash Buffer 2 to the spin column. Centrifuge for 15 sec at 8,000 g at room temperature. Discard flow-through and place column back into the collection tube. |
| 9c. | Wash and dry | Add 500 μl 80% ethanol to the spin column. Centrifuge for 2 min at 8,000 g at room temperature to dry the membrane completely. |
| 10. | Elute | Place the spin column in a new low bind collection tube (1.5 ml). Centrifuge with open lid for 5 min at 12,000 g at room temperature. Add 30 μl RNase free water directly onto the membrane of the spin column. Incubate for 1 min at room temperature. Close the lid and centrifuge for 1 min at 12,000 g at room temperature. |
| 11. | Storage | Store the RNA sample immediately at -80°C or proceed to cDNA synthesis (Step 13). |
cDNA Synthesis
Keep samples, reagents and reactions on ice (or at 4°C) at all time.
| 12. | Thaw total RNA | Thaw total RNA (from step 11) on ice. | ||||||||||||
| 13. | Prepare reagents | Thaw 5 x RT reaction buffer and nuclease free water and put on ice. Immediately before use, remove the enzyme mix from the freezer, mix by flicking the tube and place on ice. Spin down all reagents. |
||||||||||||
| 14. | Prepare cDNA synthesis mix |
If performing cDNA synthesis on multiple RNA samples, prepare a mastermix including 1 extra reaction.
Pipette 2 µl RNA template in each tube and add 8 µl cDNA mastermix. Mix by pipetting and spin down. |
||||||||||||
| 15. | Incubate and heat inactivate | Incubate the reaction at 42°C for 60 min. Heat-inactivate the reverse transcriptase at 95°C for 5 min. Immediately cool to 12°C. | ||||||||||||
| 16. | Storage | Transfer the undiluted cDNA into nuclease-free low bind tubes and freeze at -20°C for up to 5 weeks. |
qPCR Amplification
The osteomiR® test plate contains 19 different miRNA or QC primer sets. Four samples can be measured on one 96-well plate. Three columns are used per sample. 16 samples can be measured on one 384-well plate, using one row per sample.
| 17. | Thaw reagents | Thaw cDNA (from step 16) and miGreen master mix on ice for 15–20 minutes. Keep reagents on ice all the time. Before use mix the Master mix by pipetting up and down. |
Note: If you using an ABI cycler please skip to stepa 18a-20a.
| 18. | Mix cDNA with water and qPCR Master mix | Mix 2.6 μL cDNA with 127.4 μl nuclease free water, then add 130 μl miGreen master mix (in total 260 μl). Mix by pipetting up and down, spin down to collect the liquid at the bottom. Repeat this step for all samples. | ||||||||
| 19. | Prepare osteomiR™ plate | Add 10 μl reaction mixture (from step 18) (cDNA, NFW, master mix) to each of the 24 wells. Seal the plate with the appropriate optical sealing. Incubate at 4°C for a minimum of 1 hour. Note: The plate can be stored up to 16 hours at 4°C protected from light. |
||||||||
| 20. | Perform qPCR |
Before running the qPCR, spin plate for 1,000 g for 90sec. Perform qPCR and melting curve analysis as shown below. Settings have been optimized for the Roche Light Cycler® 480 II instruments.
|
If using an Applied Biosystems Instrument, following step must be adapted:
| 18a. | Mix cDNA with water and qPCR Master mix |
ROX dye is required at the following concentrations: |
|||||||||||||||
| 19a. | Prepare osteomiR® plate | Centrifuge the osteomiR® 96-well plate at 1,500 g for 90 sec, then remove the seal. Add 10 μL reaction mixture (from step 18.a) (cDNA, NFW, ROX, master mix) to each of the 24 wells. Seal the plate with the appropriate optical sealing. Incubate at 4°C for a minimum of 1 hour. Note: The plate can be stored up to 16 hours at 4°C protected from light. |
|||||||||||||||
| 20a. | Perform qPCR |
Before running the qPCR, spin plate at 1,000 g for 90 sec. Perform qPCR and melting curve analysis as shown below.” |
Data Analysis
A data analysis application (osteomiR® app) is available for all our customers. Download links will be provided upon purchase of our kits.
Troubleshooting
RNA Isolation
Poor RNA Recovery
| Column has become clogged | In most cases this can happen when recommended amounts of starting materials were exceeded. For most biofluids this is unlikely to occur. However, because of the variety of biological samples the amount of starting material may need to be decreased below the recommended levels if the column shows signs of clogging. See also “Clogged Column” below. |
| An alternative elution solution was used | For maximum RNA recovery it is recommended to elute the RNA with the RNase-free water supplied with this kit. |
| RNA content | The RNA content in serum is low therefore the concentration measurement of the purified RNA (e.g. spectrophotometric or with fluorescent dyes) is not accurately possible. The protocol is optimized using fixed volumes. |
Clogged Column
| Temperature too low | Ensure that the centrifuge and solutions remain at room temperature (18 - 25°C) throughout the procedure. Temperatures below 15°C may result in salt precipitates that may clog the columns. If salt precipitation is present, heat the solution to 30°C until completely redissolved and let the solutions cool to room temperature before use. |
Degraded RNA
| RNase contamination | RNases may be introduced when working with the samples. Ensure that proper procedures are followed when working with RNA. Please refer to “Working with RNA” at the beginning of this manual |
| Procedure not performed quickly enough | In order to maintain the integrity of the RNA, it is important that the procedure be performed quickly. |
| Improper storage of the purified RNA |
For short term storage RNA samples may be stored at -20°C for a few days. It is recommended that samples be stored at -70°C for longer term storage Tip! If possible, snap freeze your RNA in liquid nitrogen before storage in the freezer. Avoid repeated freeze/ thaw-cycles by freezing aliquots of your RNA. |
| Enzymes used may not be RNase-free | In order to prevent possible problems with RNA degradation ensure that enzymes used upstream of the isolation process are RNase-free. |
RNA does not perform well in downstream applications
| Salt or ethanol carryover | Traces of salt and ethanol from the binding step can interfere with downstream applications. Therefore, Step6 (Wash) is important for the quality of your RNA sample To avoid remaining salts please make sure that the RNA bound to the column is washed 3 times with the provided Wash Solution and ensure that the dry spin is performed, in order to remove traces of ethanol prior to elution. |
| Inhibitors | Some individual serum samples can contain inhibitors. Using spike-ins that control every step of the protocol inhibitors can be easily detected. Samples that contain inhibitors must be excluded from the analysis. |
cDNA and qPCR Amplification
| No fluorescent signal is detected during the PCR | Confirm that the PCR setup was correct by checking the signal obtained for the qPCR spike-in control „UniSp3 IPC“. |
| No fluorescent signal detected during the PCR, but the spike-in „UniSp3 IPC“ gives a valid signal. | Check that the filter in the qPCR cycler was set to either miGreen or FAM/FITC Check that the optical read is at the correct step of the qPCR cycles. |
Frequently Asked Questions (FAQ)
- How many samples can we measure with one osteomiR® kit? One kit provides sufficient reagents for the analysis of 48 serum or plasma samples. Upon request, we can provide smaller kits for proof-of-concept studies involving 10 samples or less.
- Can we use rodent or other non-human samples for the kits?
The osteomiR® tests are intended to be used with human samples.
However, orthologs for osteomiRs do exist in other species, allowing the use of adapted version of the tests in non-mammalian species. To assess the feasibility of such projects, please contact us before using the assay in non-human species. - Correlations of osteomiRs to other bone parameters? We have performed an in-depth analysis of the correlations of osteomiRs to parameters of dynamic histomorphometry and nanoCT. The results from this analysis have been presented during ECTS 2017 and have been submitted for publication.
- Does the osteomiR® test require a specific instrument or can it be run with any qPCR instrument? We recommend using the Roche LightCycler® 480. Other qPCR instruments can be used with the kit, but the software currently supports only data generated with Roche instruments. Data generated with ABI or other platforms can be analyzed manually. We will provide support in such instances.
- Which quality controls are included in the assay? Should I include additional controls? 5 control primers to monitor assay and sample quality are included in the kit, no further controls are necessary.
- I want to analyze a sample type that is not listed as tested yet, can I do that?
Please contact info@bmgrp.com to discuss the possibilities. - What is the recommended RNA extraction method? All our RUO kits include a protocol and reagents for RNA extraction. TAmiRNA´s osteomiR® kit has been standardized to a volume of 200 µl serum as starting material to ensure high RNA yield and prevent inhibition of downstream PCR amplification using the miRCURY™ RNA isolation kit, which is included in the osteomiR® Kit. For further information read our guidelines detailing RNA purification and sample preparation for your osteomiR® kit.
- What is the sensitivity of the assay? LOD: 10 copies/µl (6 replicates, 95% positive detection); LLOQ: 35 copies/µl
- What is the reproducibility of the assay? Intra-assay (n=6) ≤ 20%, Inter-assay (n=3) ≤ 20%
- What data analysis tools do you recommend? We recommend using the software included in the kit for normalization and quality control.
 Download biomedica product list
Download biomedica product list